NIR Model Development at Celignis
Background to NIR
Near Infrared Spectroscopy (NIRS) provides a rapid means to analyse a wide variety of feedstocks. It requires minimal sample preparation, is non-destructive, and
can have a high throughput on a sample basis.
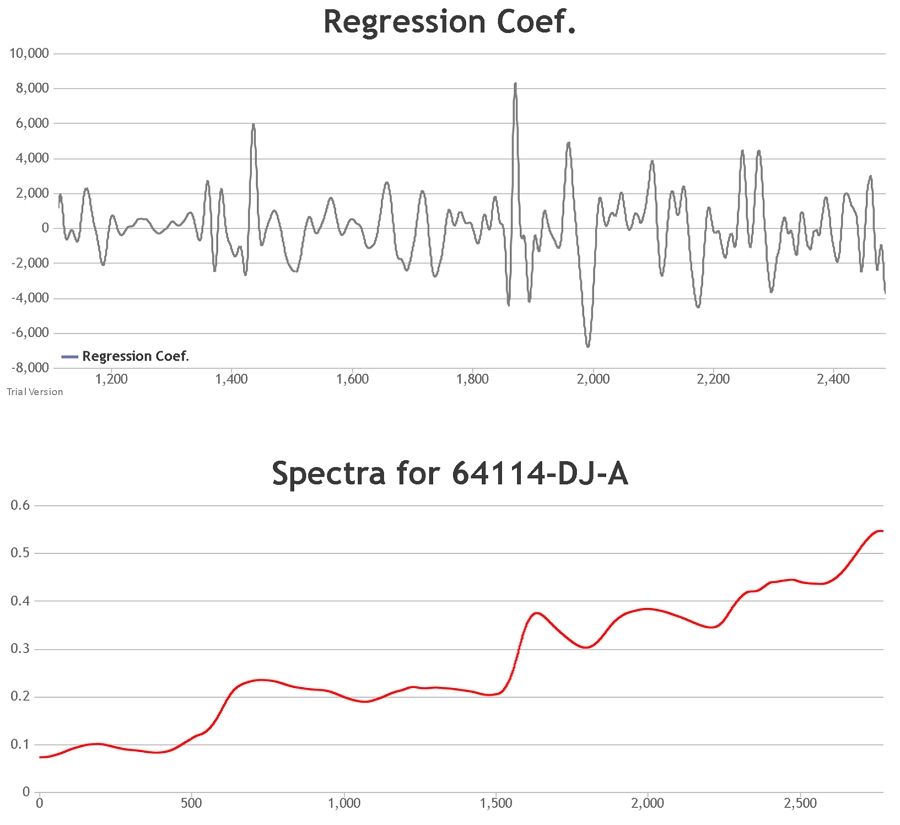
The procedure involves the focussing of radiation on a sample. While some of the radiation will be scattered, some will pass through the sample, interacting with it. When the radiation finally reaches the detector it will pass on this absorbance information. This, along with the scatter from the sample, forms the spectrum of the material.
The procedure involves the focussing of radiation on a sample. While some of the radiation will be scattered, some will pass through the sample, interacting with it. When the radiation finally reaches the detector it will pass on this absorbance information. This, along with the scatter from the sample, forms the spectrum of the material.
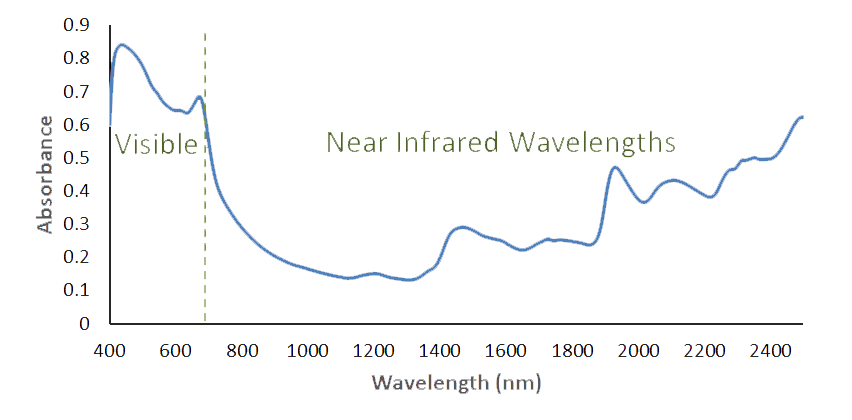
The FOSS XDS spectroscopy device used at Celignis Analytical detects radiation in the wavelength region of 400 to 2500nm. Visible light is
defined as 400 to 700nm with the longer wavelengths being in the near infrared region. The CH, OH and NH bonds of an organic substance (those of most interest in carbohydrate
chemistry) will absorb energy in this region. The infrared spectrum consists of overtones and combination bands of these and other fundamental absorptions.
NIR Model Development
NIRS is largely an indirect analytical technique requiring calibration using samples of known composition determined by using standard, wet-chemical,
methods. These calibrations are based on the correlations of spectra of samples with their wet chemistry data. Once calibrated,
quantitative predictions for the composition of a sample can be attempted from its NIR spectrum alone. The results and accuracy of
calibrations are then validated by presenting "unknown" samples to the NIR device and comparing the results with those obtained by the wet-chemistry analysis.
At Celignis spectra are collected using a FOSS XDS Spectrophotometer and the Vision software program. The spectra are then imported into our custom-built chemometric software program for subsequent treatment and model development. This software was developed by Celignis in the BIOrescue BBI project. Partial least squares regression using one Y variable (i.e. PLS1) is used for the development of Celignis models.
At Celignis spectra are collected using a FOSS XDS Spectrophotometer and the Vision software program. The spectra are then imported into our custom-built chemometric software program for subsequent treatment and model development. This software was developed by Celignis in the BIOrescue BBI project. Partial least squares regression using one Y variable (i.e. PLS1) is used for the development of Celignis models.
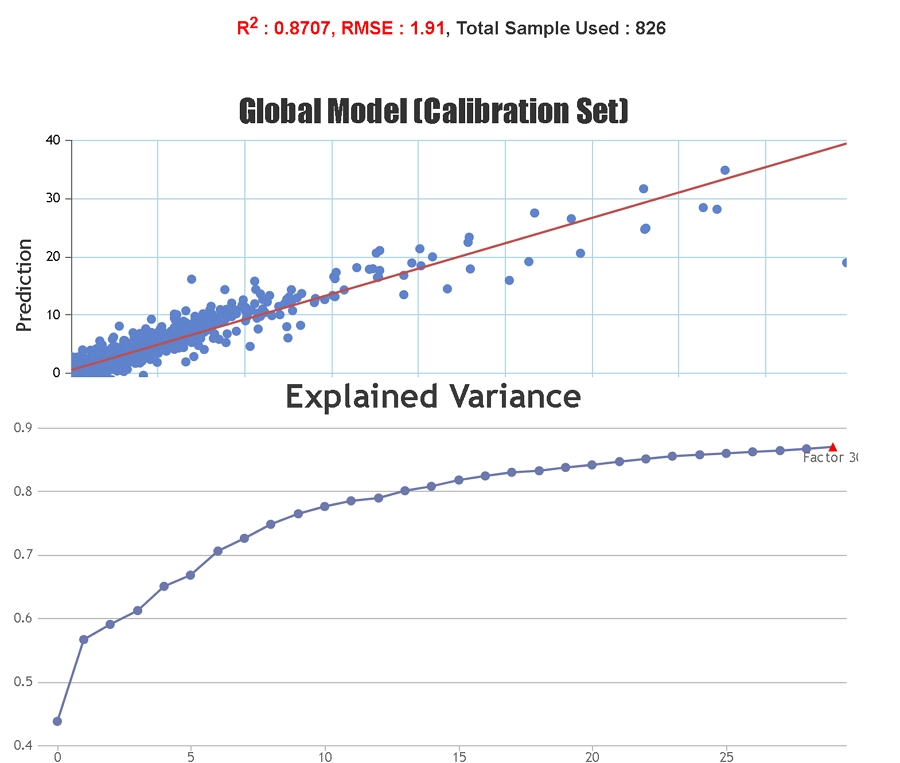
Spectral pre-treatment techniques are often applied prior to model development in order to simplify the models and reduce the effect that particle size variation may have on the scattering of light. At Celignis PLS models have been tested on the raw spectra and on spectra treated with various techniques including: multiplicative scatter correction (MSC), extended multiplicative scatter correction (EMSC), standard normal variate (SNV), standard normal variate and detrend (SNVDT), and Savitzky-Golay (SG) derivatives (1st to 4th order). Cross validation statistics have been used to determine the most appropriate model. The Haaland and Thomas (1988) criterion is used to select the number of PLS factors to use in the model.
A good model for the prediction of unknown samples should cover a wide variety of sample types and compositional values so that the unknown sample should not be a spectral/chemical/physical "outlier" but instead should be of a similar spectral and physico-chemical composition to some of the samples used to build the model.
A good model for the prediction of unknown samples should cover a wide variety of sample types and compositional values so that the unknown sample should not be a spectral/chemical/physical "outlier" but instead should be of a similar spectral and physico-chemical composition to some of the samples used to build the model.
Important Statistics
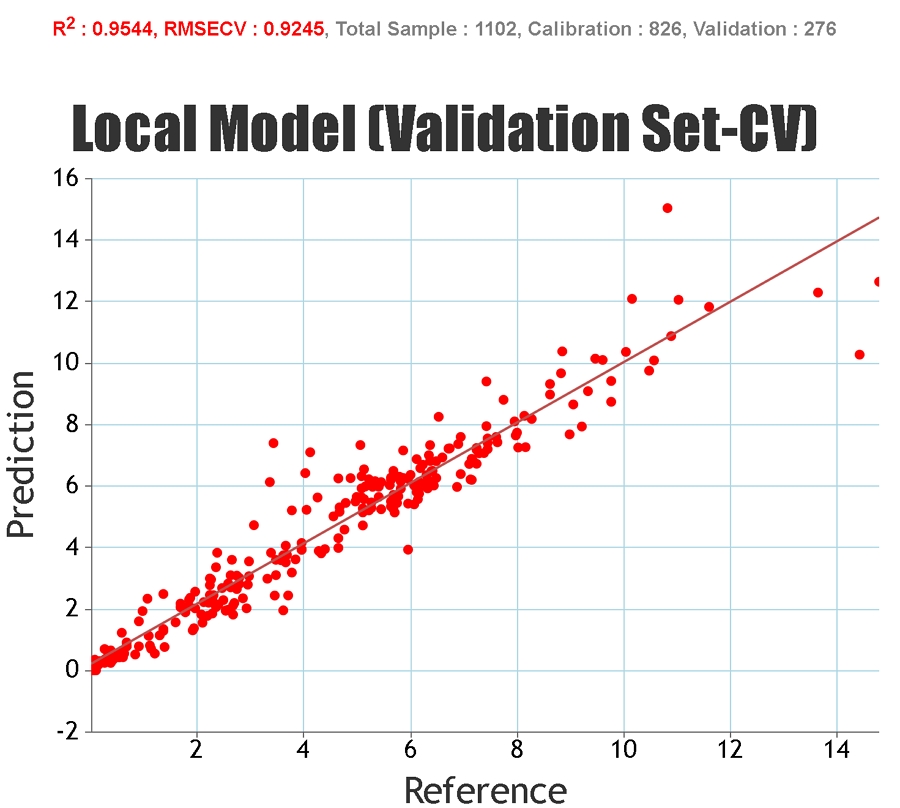
In order to get an idea of the predictive ability of a model a number of statistical measures are used. These can be applied to the
calibration set (the group of samples that are used to build the model parameters), the cross-validation set (samples temporarily
excluded from model development but still ultimately involved in the development of the model), and the independent set (samples
that have no input into the development of the model).
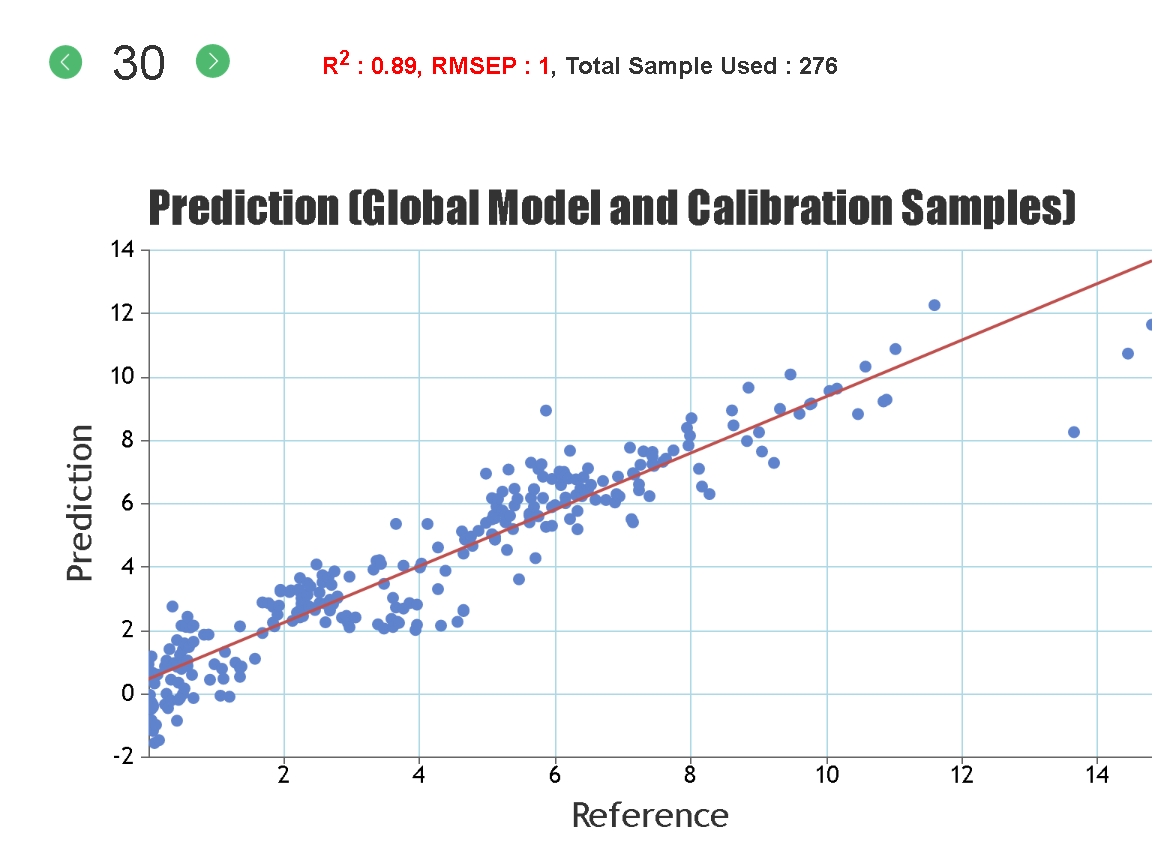
Statistics solely based on the calibration set can give an inaccurate representation of the predictive ability of the model for unknown samples since it is possible to "overfit" the model to the calibration set, particularly if a large number of PLS factors are used. Cross-validation statistics provide a better idea of the robustness of a model but, ideally, independent validation (a test set) should be used. When presenting our regression statistics Celignis will use the values for the test set, unless otherwise stated.
Some of the most important statistics, those that are used on this website, are described below:
Statistics solely based on the calibration set can give an inaccurate representation of the predictive ability of the model for unknown samples since it is possible to "overfit" the model to the calibration set, particularly if a large number of PLS factors are used. Cross-validation statistics provide a better idea of the robustness of a model but, ideally, independent validation (a test set) should be used. When presenting our regression statistics Celignis will use the values for the test set, unless otherwise stated.
Some of the most important statistics, those that are used on this website, are described below:
R-Squared, Coefficient of Multiple Determination - Describes how well the data points fit the statstical model (the line of regression). Values range from 0 to 1. A 100% accurate model would have an R-Squared of 1 with all samples lying on the regression line.
RMSEP, Root Mean Square Error of Prediction - This measures the average accuracy (i.e. the difference between the true and estimated compositional value) of the prediction. For the samples in the test set it can be considered that 2 times the RMSEP represents a 95% confidence interval for the real compositional value. For example, if the model predicts a glucan content of 40% and the RMSEP is 1%, then there is a 95% chance that the glucan content of that sample, as measured by wet-chemical means, lies between 38 and 42%.
Bias - This is defined as the average difference between the NIR-predicted value and the real value. A positive value means that, on average, the model is over-estimating the composition by this amount whilst a negative value represents an underestimation.
RMSEP, Root Mean Square Error of Prediction - This measures the average accuracy (i.e. the difference between the true and estimated compositional value) of the prediction. For the samples in the test set it can be considered that 2 times the RMSEP represents a 95% confidence interval for the real compositional value. For example, if the model predicts a glucan content of 40% and the RMSEP is 1%, then there is a 95% chance that the glucan content of that sample, as measured by wet-chemical means, lies between 38 and 42%.
Bias - This is defined as the average difference between the NIR-predicted value and the real value. A positive value means that, on average, the model is over-estimating the composition by this amount whilst a negative value represents an underestimation.
SEP, Standard Error of Prediction - Whilst the RMSEP measures the accuracy of prediction,
the SEP measures the precision of the prediction (i.e. the difference between repeated measurements). The SEP squared is approximately equal to the RMSEP squared minus
the Bias squared. Hence, if the bias is low the values for RMSEP and SEP will be similar. Since we at Celignis are focused on getting predictions to be as accurate as
possible, we consider the RMSEP to be more important.
RPD, Ratio of standard error of Performance to standard Deviation - This is equal to the SEP divided by the standard deviation of the compositional values (determined via wet-chemistry) of the samples in the test set. Whilst the RMSEP, SEP, and Bias use the same units of measurement as the constituent (e.g. percent for glucan content), the R-Squared, RPD, and RER (see below) values are dimensionless, meaning that they can be compared on the same basis between models for different constituents/properties. If the RPD is equal to one then the SEP is equal to the standard deviation of the reference data meaning that the model is not predicting the reference values. Higher values for the RPD suggest increasingly accurate models.
RPD, Ratio of standard error of Performance to standard Deviation - This is equal to the SEP divided by the standard deviation of the compositional values (determined via wet-chemistry) of the samples in the test set. Whilst the RMSEP, SEP, and Bias use the same units of measurement as the constituent (e.g. percent for glucan content), the R-Squared, RPD, and RER (see below) values are dimensionless, meaning that they can be compared on the same basis between models for different constituents/properties. If the RPD is equal to one then the SEP is equal to the standard deviation of the reference data meaning that the model is not predicting the reference values. Higher values for the RPD suggest increasingly accurate models.
RER, Range Error Ratio - This is equal to the range in the compositional values
(i.e. the maximum value minus the minimum value) divided by the SEP. The numbers obtained for the RER will typically be around four to five times larger than those for the RPD;
however, the exact relationship between the two will depend on the distribution of samples in the test set. AACC Method 39-00.01 (1999) provides quality thresholds for model
performance based on the RER values: For an RER > 4 - the calibration is acceptable for sample screening; for an RER > 10 - the calibration is acceptable for quality control;
and for an RER > 15 - the calibration is good for quantification. Thresholds are also provided for the RPD value but this value can be subject to manipulation according to
how the sample set is constructed. Celignis considers that the RER value is a better test for the quality of the model, providing that there are no concentration outliers to
inflate the value and that the concentration range of the constituent is well represented (as is the case in the Celignis NIR models).
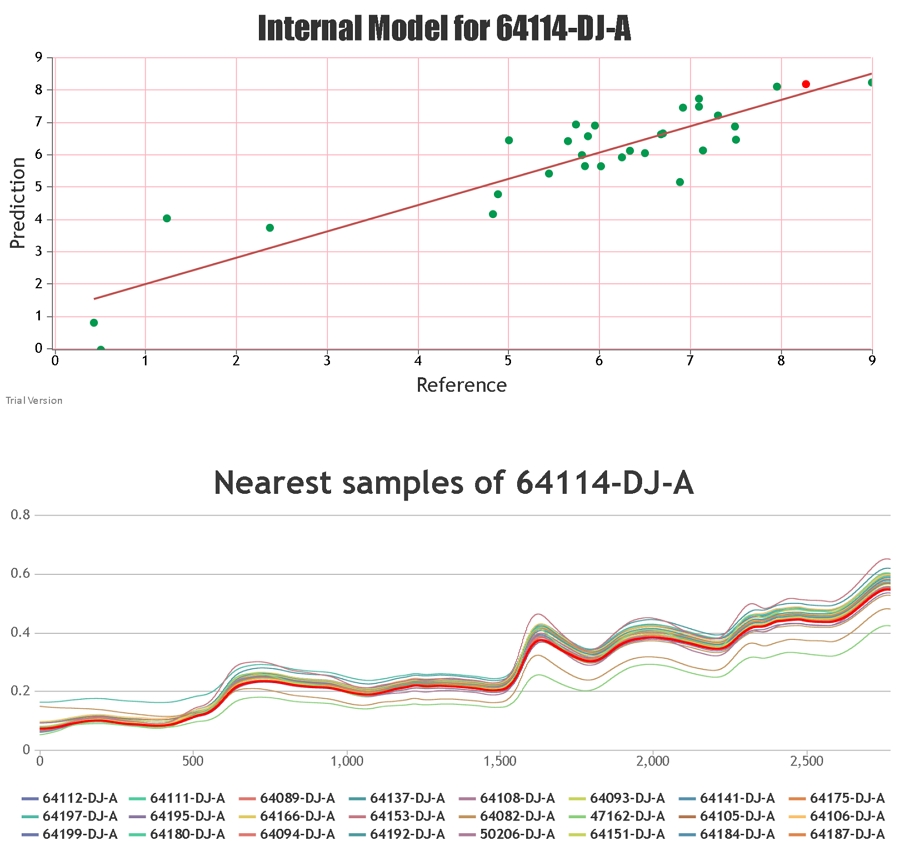
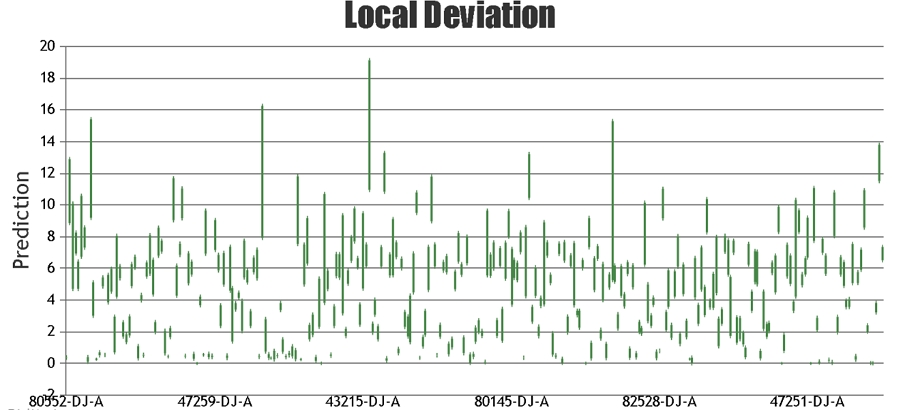
When providing the results for NIR predictions of samples, Celignis provides the predicted compositional value and also a value for the "Deviation in Prediction". This Deviation value
can be considered to represent a form of a confidence interval around the predicted value. It is calculated based on how similar the spectrum of the
unknown sample is to the samples that constitute the calibration set.
If the type of sample to be predicted is already in the calibration set then the Deviation is likely to be low. Click here for a list of some of the sample types in the current Celignis models.
If the type of sample to be predicted is already in the calibration set then the Deviation is likely to be low. Click here for a list of some of the sample types in the current Celignis models.
A sample with a large value for its deviation will be quite different spectrally, and quite possibly phsically and chemically, from the calibration samples, hence the model
may not be appropriate for predicting that sample with a high degree of accuracy.
At Celignis, we pride ourselves on the accuracy and precision of our analysis. Customer satisfaction is of paramount importance. For our NIR Analysis Package, if we find that the deviation in prediction is relatively high (defined as a value over 3% for the total lignocellulosic sugars content) then we will undertake the chemical analysis of that sample at no extra charge and provide you with all of the data that we obtain in this analysis. This chemical analysis will cover the analysis packages that we used to develop our NIR models: P3 - Ash Content, P4 - Ethanol Extractives, and P9 - Lignocellulosic Sugars and Lignin.
Click here for more information on the NIR models that have been developed by Celignis and here for some of our peer-reviewed publications on them.
At Celignis, we pride ourselves on the accuracy and precision of our analysis. Customer satisfaction is of paramount importance. For our NIR Analysis Package, if we find that the deviation in prediction is relatively high (defined as a value over 3% for the total lignocellulosic sugars content) then we will undertake the chemical analysis of that sample at no extra charge and provide you with all of the data that we obtain in this analysis. This chemical analysis will cover the analysis packages that we used to develop our NIR models: P3 - Ash Content, P4 - Ethanol Extractives, and P9 - Lignocellulosic Sugars and Lignin.
Click here for more information on the NIR models that have been developed by Celignis and here for some of our peer-reviewed publications on them.